A Focused Research Organization to Measure Complete Neuronal Input-Output Functions
Measuring how neurons integrate their inputs and respond to them is key to understanding the impressive and complex behavior of humans and animals. However, a complete measurement of neuronal Input-Output Functions (IOFs) has not been achieved in any animal. Undertaking the complete measurement of IOFs in the model system C. elegans could refine critical methods and discover principles that will generalize across neuroscience.
Systems neuroscience aims to understand the complex interplay of neurons in the brain, which enables the impressive behaviors of animals. A critical component of this understanding is the Input-Output Function (IOF) of neurons, which characterizes how a neuron integrates its inputs and responds to them. However, despite its importance, a complete measurement of IOFs in any animal has not been achieved, creating a significant blind spot in our understanding of brain function. While parts of IOFs have been measured through various experiments, these efforts only control a narrow subset of the inputs to any given neuron, providing only a small slice of the true IOF. Furthermore, the output of neurons is a complex nonlinear function of their inputs, adding to the complexity of the problem. To truly understand the computation and function of the brain, we need a detailed functioning of the IOFs, which requires controlling all of the inputs and observing the factors that shape IOFs. Pursuing this in a single animal model would uncover new methods, tools, and scientific principles that could catalyze large-scale innovation across neuroscience.
Project Concept
The project aims to unlock a deeper understanding of how neurons in the brain process information through the complete mapping of neuronal IOFs using the model organism C. elegans. This comprehensive mapping is a vital step in understanding how the brain’s neurons receive, integrate, and respond to signals. The project will use a combination of advanced techniques including optogenetics, modern microscopy, and microfluidics to control and observe the nervous system of the worms, and will develop models to predict how neurons respond based on their inputs. The team will also explore how different factors, such as chemicals, drugs, and non-neural cells influence these responses. To ensure maximal benefit to the field and the scientific community, all data and findings will be shared openly and resources will be allocated to promoting collaboration with outside experts on experimental design, technology development, and computational and theoretical analysis.
What is a Focused Research Organization?
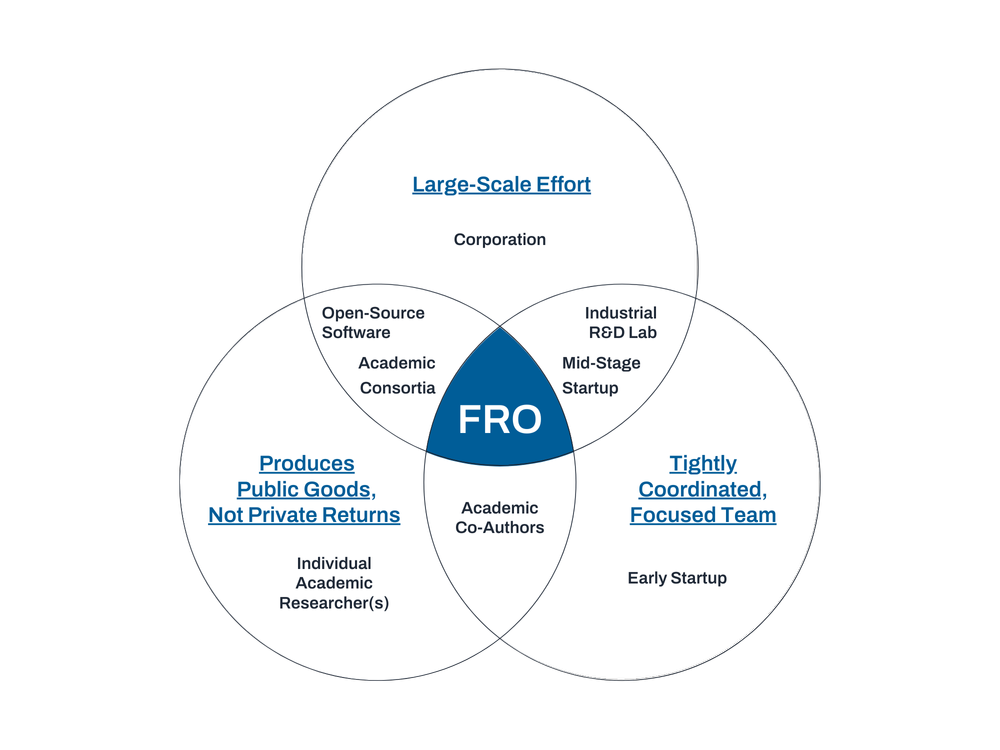
Focused Research Organizations (FROs) are time-limited mission-focused research teams organized like a startup to tackle a specific mid-scale science or technology challenge. FRO projects seek to produce transformative new tools, technologies, processes, or datasets that serve as public goods, creating new capabilities for the research community with the goal of accelerating scientific and technological progress more broadly. Crucially, FRO projects are those that often fall between the cracks left by existing research funding sources due to conflicting incentives, processes, mission, or culture. There are likely a large range of project concepts for which agencies could leverage FRO-style entities to achieve their mission and advance scientific progress.
This project is suited for a FRO-style approach because it requires a level of coordinated development and engineering that is too big for a single academic lab, too complex for a loose multi-lab collaboration, and not directly profitable enough for a venture-backed startup or industrial R&D project. The project also aims to create a suite of public goods through its commitment to open science, with plans to share data and code as they are developed. More broadly, the work lends itself well to the development of a new set of tools and methodologies rather than the products or papers incentivized by traditional research models.
How This Project Will Benefit Scientific Progress
This project aims to revolutionize neuroscience by providing the first complete measurement of neuronal Input-Output Functions (IOFs) in the model organism C. elegans. Identifying causal interactions between neurons will provide unprecedented insight into brain function, and the methods developed in the process pave the way for understanding more complex nervous systems.
Key Contacts
Authors
- Konrad Kording, University of Pennsylvania, koerding@gmail.com
Referrers
- Jordan Dworkin, Federation of American Scientists, jdworkin@fas.org
A Focused Research Organization to Develop a Modular and Scalable Platform for Human Molecular Monitoring
Wearable health electronics are now ubiquitous, but continuous molecular monitoring is only widely available for glucose. Decades of research have expanded continuous monitoring to other molecules, but these techniques are restricted to research labs and remain disconnected from daily human use. We propose a platform to translate and distribute these emerging technologies, enabling the mapping of the time-varying human metabolome and the design of closed-loop devices for personalized health.
Humans are the best model organisms for humans, yet we have few tools to study human biochemistry in situ and in real time. The Human Metabolome Database lists ~20,000 detected compounds, of which only ~3,000 have been quantified. Even fewer of these biomolecules have been studied with time resolution in longitudinal human studies.
Cardiovascular, metabolic/endocrine and drug pharmacokinetic phenomena are driven by biomolecules varying on the time scale of seconds to minutes to hours, impacting behavior and well-being on similar time scales. Currently, the only way to measure these molecules is through laboratory testing, which is obtrusive to daily life. Moreover, laboratory testing cannot be conducted at the frequency necessary to capture all of these variations.
In contrast, wearable monitors are user-friendly and enable continuous measurements at the correct time scale. These monitors require specially engineered biosensors, since there are a limited number of naturally-occurring enzymes that generate continuous, time-varying electrical signals like those used by continuous glucose monitors, which are currently the only commercially available device of this kind. However, most labs that pioneer biosensing strategies do not develop human-compatible devices and vice versa, creating a chasm between these two areas of research and development.
Project Concept
This project aims to achieve minimally invasive continuous monitoring of 100+ analytes in the human body and deliver devices to researchers. This project will develop a medical device testbed that can use the myriad biosensing strategies pursued by academic labs to develop devices for human experiments. Our technical approach combines synthetic ion channels and conformational switches coupled with a CMOS array to assess many analytes in parallel. We already have access to customized fabrication techniques, and the cost and barriers in designing and manufacturing proteins and silicon sensors are trending downwards.
The project will progress through four interdependent stages:
- Survey and prioritize metabolic, hormone, and immune targets that provide the greatest explanatory power for well-being. Select biosensor and transduction systems reported in the literature to interface with our platform.
- Develop an integrated circuit functionalized with biosensors for parallel multi-analyte sensing packaged in a wearable form factor. Test devices in humans to validate against conventional blood sample analyses.
- Specify and share validated devices in bulk to catalyze large-scale human field research.
- Curate a time-varying human metabolome.
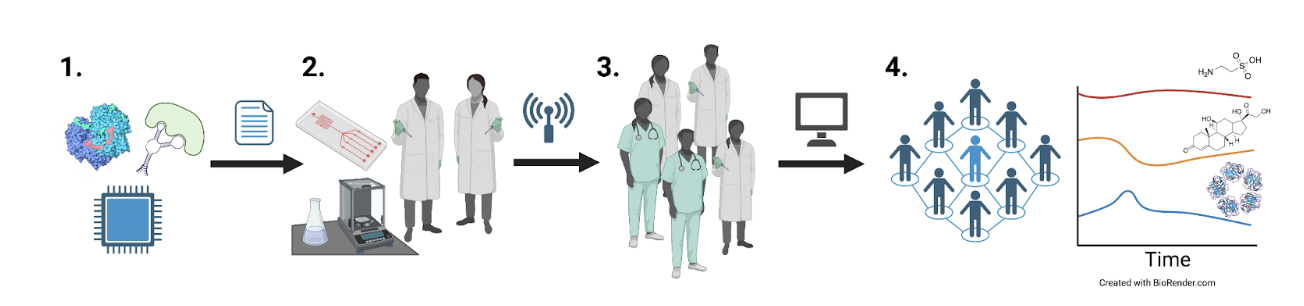
The project will progress through four interdependent stages.
What is a Focused Research Organization?
Focused Research Organizations (FROs) are time-limited mission-focused research teams organized like a startup to tackle a specific mid-scale science or technology challenge. FRO projects seek to produce transformative new tools, technologies, processes, or datasets that serve as public goods, creating new capabilities for the research community with the goal of accelerating scientific and technological progress more broadly. Crucially, FRO projects are those that often fall between the cracks left by existing research funding sources due to conflicting incentives, processes, mission, or culture. There are likely a large range of project concepts for which agencies could leverage FRO-style entities to achieve their mission and advance scientific progress.
This project suits a FRO-style model because the four research stages require tight feedback loops and standardization and the medical device industry is not incentivized to pursue this kind of research. Private companies typically focus on a handful of molecules most relevant to diabetes care, taking advantage of proven biosensors and predictable insurance reimbursement. A stand-alone, non-profit institute is best suited to standardize and derisk experiments on molecular monitoring to catalyze the formation of a consortium of experimenters. Just as the nonprofit AddGene has standardized and democratized access to genetic material, we seek to develop the analog institution for medical devices.
How This Project Will Benefit Scientific Progress
Human physiology is currently a poorly explored, high-dimensional space, and scientific labs lack the tools to measure human biochemistry over time. The devices developed through this project will first help scientists study specific questions in domains such as disease etiology, human behavior, and drug discovery. Study validity will be enhanced by providing additional molecular time courses which will clarify the relationships between conditions, specific biomarkers, and interventions. Next, these studies will enable the development of a human molecular atlas, similar to the Human Metabolome Database but with time resolution. This database could act as a powerful tool for developing nuanced models of human physiology to uncover previously overlooked phenomena. Finally, similar devices will ultimately become accessible as consumer health products, enabling the next generation of personalized health.
Key Contacts
Authors
- Anand K. Muthusamy (amuthusa@caltech.edu) and
- David Garrett (dgarrett@caltech.edu), Caltech
Referrers
- Alice Wu, Federation of American Scientists, awu@fas.org
Learn more about FROs, and see our full library of FRO project proposals here.
The overarching strategy involves converting biochemical interactions into a time-varying electrical signal and isolating specific interactions into separate channels on a CMOS chip. We envision a universal platform that can accept the myriad of biosensors from labs worldwide. Advances in synthetic biology and protein engineering have unlocked various modes of biosensing. Oxidoreductases, like glucose oxidase used in CGMs, provide a direct conversion of the local ligand concentration to an electrical current. Recent advances in deep learning protein engineering have shown proof of concept for the de novo design of enzymes, but some classes of biomolecules such as nucleic acids and proteins would be out of reach for enzymatic detection. In turn, we can either mimic nature’s ion channels or create our own electronic transduction systems. Aptamers are amenable to high-throughput evolution to bind a variety of targets and provide a hairpin motion that can move redox probes to and away from an electrode surface (cf. example of a cortisol sensor). Recent advances in DNA origami have enabled synthetic, self-assembling nanopores whose current can be modulated by the binding of other biomolecules. Both biological and solid-state nanopores are increasingly used in commercial scientific equipment, providing a tailwind for medical devices.
All of these biosensor and transduction systems can be integrated into field-effect transistors (FETs). Even strategies involving membrane proteins can now be interfaced with transistors thanks to organic electrochemical transistors with supported lipid bilayers harboring ion channels and nanopores. Modern complementary metal oxide semiconductor (CMOS) processes allow us to parallelize many sensors onto a single device at a low per-unit cost. Each sensor consists of a FET sensing unit, where the presence of molecules of interest induces a surface potential in the FET gate. This modulates the current in the FET, which is then measured with read-out circuitry. Each sensor’s current is then digitized and used as an indicator of molecular concentrations. Since relatively few channels are required (~100s) compared to conventional CMOS devices, coarser fabrication processes could be used (e.g. 130 nm) which would reduce cost and time while still allowing for a compact active area (~mm2).
The CMOS device will be packaged in a wearable form factor miniaturized for on-body measurements using a battery and wireless data transmission to a smartphone (e.g. using Bluetooth low-energy). We will draw interstitial fluid via reverse iontophoresis to reach the CMOS chip sitting next to the skin surface. Since different analytes have different rates of variation and concentration ranges, we will model the appropriate temporal resolution and SNR we can achieve. This device form factor and reverse iontophoresis method has been validated for ~mM glucose measurements with ~min resolution. In contrast, hormones may be present at ~6 orders of magnitude lower concentrations but only vary over days-weeks. Using our CMOS configuration, different channels can be read out individually. To efficiently capture these changes, different sampling rates will be used to ensure we capture their peaks and troughs while minimizing energy consumption. For analytes with lower baseline concentrations, techniques like averaging (over multiple samples and CMOS channels) may be required to improve sensitivity. The mean absolute relative difference (MARD) will be computed against blood LC-MS ground truth.
We are in a position where our data science methods outpace quality datasets. For example, geometric deep learning is useful for mapping interactions in a metabolome, and long short-term memory is well-established for analyzing and predicting time series data. These data can become actionable in a closed-loop device where a biosensing event triggers an alert or pharmacological intervention as in an artificial pancreas. Here again, control theory approaches to physiology are sufficiently mature.
Focused Research Organizations: A New Model for Scientific Research
Scientific research and development often occurs in starts and stops: insights and innovations grow rapidly out of new research fields, but over time, the low hanging fruit is picked off, bottlenecks arise, and progress dwindles. Overcoming these bottlenecks can be extremely rewarding because of the potential to trigger a cascade of follow-on research. Yet, university labs and commercial organizations are not always incentivized to tackle these challenges. Academic incentives disfavor medium- to large-scale teamwork across disciplines and projects with a low possibility of publishable results, while commercial profit motive precludes the production of public goods, so projects that fall in between academic and commercial incentives often go untouched.
Enter Focused Research Organizations (FROs), a new type of non-profit research organization designed for this middle ground. FROs are structured like start-ups with tightly coordinated, medium-sized teams that allow them to engineer and scale solutions in a way for which university labs are ill-suited. FROs focus on solving well-defined challenges to produce public goods that are neither profitable nor publishable: processes, tools, and datasets that enable the use of new research methods and accelerate the pace of scientific research. Successful large-scale research collaborations of the past like the Large Hadron Collider and the Human Genome Project could be considered similar such projects since they produced tools and datasets that enabled new particle physics and genomics research, though they’re of a far larger size and scope. While we may only fund one or two such large scale projects in each generation, there are plenty of mid-scale problems that could be economically solved with a FRO.
Launched in 2021, Convergent Research is a non-profit organization dedicated to incubating and funding new FROs, and their work is growing: in March of this year, Forbes reported that Convergent Research received $50 million in philanthropic commitments to launch two new FROs, doubling the size of their small portfolio. Convergent Research FROs are now:
- developing tools to map all of the neural circuits in the brain and advance neuroscience,
- identifying the unintended/unknown molecular targets of every approved pharmaceutical drug to improve drug development and repurposing,
- developing technologies to scale protein analysis by 100x and accelerate proteome biology research,
- and creating open source tools for life scientists to conduct synthetic biology research on a more diverse set of microorganisms.
While Convergent Research has so far only funded FROs working in the life sciences, this research model can be applied to bottleneck research problems in any field.
Convergent is also not the only organization experimenting with this model though: the UK government’s new Advanced Research and Invention Agency (ARIA), inspired by Advanced Research Projects Agencies (ARPAs) in the U.S., plans to fund FROs as part of its strategy. ARIA will be the first governmental program to fund FROs, and hopefully not the last. Philanthropic funding simply cannot compare with the scale of government funding for scientific research. As the largest science funder in the world, the U.S. federal government provides enormous potential to expand the number of FROs that get launched over the next decade to accelerate scientific progress.
To help agencies conceptualize the types of scientific and engineering problems that FROs can address, FAS and Convergent Research have come together to create a database of vetted FRO proposals from researchers across the sciences. These proposals tackle a range of problems from developing neuromorphic AI hardware using superconducting optoelectronic networks to building open-source computational infrastructure for the monitoring, reporting, and verification of ocean-based carbon removal. Curious agencies can look to our database for inspiration on how FROs can fit into their particular research priorities. If any agency finds a proposal particularly compelling, we welcome them to reach out to FAS or to the proposal authors for more details.
As federal agencies begin developing their FY25 budget proposals this month, those that fund scientific research—the National Science Foundation, the National Institutes of Health, the Department of Energy (DOE), the National Aeronautics and Space Administration, the Department of Defense, the Department of Agriculture (USDA), and the National Oceanic and Atmospheric Administration (NOAA)—should follow in the UK’s footsteps and request pilot funding for FROs to complement their more traditional research programs. Because of their potential to unlock bottlenecks and improve overall research productivity, FROs can be a means of enabling more scientific progress with less funding under the new budget deal.
Beyond agency interest and funding, Congressional authorization can also help launch FROs within the U.S. government. All of the agencies mentioned above have the Other Transactions Authority (OTA) necessary to pilot FROs, except for the USDA and NOAA. (AgARDA within USDA does have OTA, but it currently lacks funding.) Thus, Congress should prioritize providing the USDA and NOAA with OTA for R&D such that the agencies can not only experiment with FROs, but also other novel research models that are useful for their mission but are not traditional grants or contracts. For all science agencies, explicit Congressional authorization to fund FROs would motivate and assure agencies of their legal authority to fund FROs, since some agencies, like the DOE, have been historically more conservative in their use of OTA than others.
A Focused Research Organization to Systematically Study Bacteriophage Genes and their Functions
Systematically sequencing the genome and studying the function of genes from all viruses that infect a set of model bacteria with significant scientific, biotechnological, and human health relevance will enable the development of phage-gene libraries that can in turn enable the faster development of genetic tools for advancing molecular biology.
Problem Statement
Viruses have been evolving host-modifying factors for billions of years. This wealth of naturally engineered proteins holds the key to unlocking the full potential of the cell. Virus-derived genetic tools have driven most key advances in molecular biology, from recombinant DNA to CRISPR genetic engineering. Although most such transformative discoveries have resulted from the study of bacteriophages (viruses of bacteria), phage research has relied primarily on inferential work rather than systematic approaches to discern the functions of phage genes. With serendipity as the primary engine of discovery, experimental approaches have not kept pace with phage genome sequencing over the past decade. Consequently, the vast majority of phage genetic diversity is still entirely unexplored.
Project Concept
We have built a high throughput screening platform to characterize phage genes and completed a pilot of the entire pipeline, from gene selection through functional screening and mechanistic follow-up (manuscript in preparation and available upon request). Our FRO will scale this platform and use it to chemically synthesize and test all non-redundant phage genes from two clinically relevant families of bacteria (Enterobacteria and Mycobacteria) which collectively host ~40% of all isolated phages, allowing us to test a large swathe of phage genetic diversity in a set of model species. The phage-gene library generated from this process will enable us to pursue the following objectives: 1) discover new molecular tools with revolutionary potential (eg. broadly understanding the principles of protein detection in antiviral immunity could yield a generalizable protein-targeting framework without some of the pitfalls of antibodies), 2) develop therapeutic avenues for antimicrobial resistant infections inspired by natural antiviral defense and counter-defense strategies, 3) build an inventory of phage design principles and engineering methods for therapeutic, industrial, and microbiome-directed applications, and 4) gain a complete understanding of interactions between phage and their hosts.
What is a Focused Research Organization?
Focused Research Organizations (FROs) are time-limited mission-focused research teams organized like a startup to tackle a specific mid-scale science or technology challenge. FRO projects seek to produce transformative new tools, technologies, processes, or datasets that serve as public goods, creating new capabilities for the research community with the goal of accelerating scientific and technological progress more broadly. Crucially, FRO projects are those that often fall between the cracks left by existing research funding sources due to conflicting incentives, processes, mission, or culture. There are likely a large range of project concepts for which agencies could leverage FRO-style entities to achieve their mission and advance scientific progress.
This project is suited for a FRO-style approach because to achieve our scientific goals, we will need to scale our platform ~10,000-fold from 104-5 assays in the pilot to ~108-9 assays at the FRO. Massively parallelizing these assays will involve a highly systematic effort with a tightly coordinated and dedicated team, a substantial initial investment in gene-library synthesis and platform engineering, and long publishing timelines, which are qualities unsuitable for traditional grant funding. For these reasons, an FRO is the ideal (and probably the only viable) structure for this project.
How This Project Will Benefit Scientific Progress
Paradigm shifts in biology have often started with the humble bacteriophage. With 108-9 prospects across the oldest and most diverse host-pathogen interface in the biosphere, our FRO presents abundant opportunities for making impactful discoveries, and will pioneer a new field of functional metaviromics. Moreover, the phage-gene libraries we will create are analogous to small-molecule screening libraries, consisting of 104-105 phage-derived natural products that can be used to find potentiators or suppressors of any cellular stressor of interest. We expect these resources to enable discovery far beyond the scope and timeline of our FRO.
Key Contacts
Authors
- Sukrit Silas, University of California San Francisco,
Referrers
- Alice Wu, Federation of American Scientists, awu@fas.org
Learn more about FROs, and see our full library of FRO project proposals here.
A Focused Research Organization to Reduce Antibiotic Resistance In Aquaculture
Research and engineering to reverse antibiotic resistance in aquatic bacteria, through the application of a well-validated CRISPR-based genetic system, can help catalyze safer, more sustainable land-based aquaculture as a nutritious and affordable food source.
The growing human population needs affordable, healthy sources of protein. With overfishing putting severe pressure on global fish stocks, aquafarming presents a potential alternative. The U.S. currently imports about 80% of its seafood, and most imports are produced by foreign aquaculture; expanding domestic aquaculture could help to close the $17 billion seafood trade deficit. But domestic aquafarming poses its own challenges, including the potential for environmental contamination near ocean-based operations. In such scenarios, high concentrations of fish within netted areas lead to bacterial and other waste contamination spreading beyond the arena of fish confinement. The alternative strategy of raising fish in isolated inland enclosures may pose less environmental risk, but also requires maintenance of water quality, frequent water filtration and, often, the use of high antibiotic concentrations mitigate bacterial fish pathogens that thrive in such overcrowded conditions. In practice, aquafarmers often try to reduce the level of antibiotics added to the water in the last few weeks of fish growth to drop their concentrations below mandated health standards for commercial fish, but these efforts are only partly effective and create significant logistical burdens.
Project Concept
We proposed the development of genetic systems to reduce the prevalence of antibiotic resistance in land-based aquafarming enclosures. We will develop harmless strains of environmental bacteria capable of transferring self-copying genetic cassettes to pathogenic bacterial strains of concern in aquaculture. With these strains, we aim to reduce virulence of those bacterial pathogens in high-density fish enclosures and scrub their antibiotic resistance.
The heart of the project is to apply a well-validated self-amplifying genetic system, referred to as Prokaryotic-Active Genetics (Pro-AG), to the task of scrubbing virulence and antibiotic resistance factors from bacterial pathogens in aquaculture facilities. Since publication of the seminal study describing this CRISPR-based system for reversing antibiotic resistance (Valderrama et al., 2019, Nat. Comm. 10, 5726), we have further advanced the Pro-AG platform by combining it with means of spreading between bacteria through horizontal transfer systems such as conjugal transfer elements or bacteriophage. We have also incorporated new genetic features to the Pro-AG toolkit including a system to cleanly and efficiently delete genetic elements such as virulence factors responsible for antibiotic resistance. Building on these core achievements, we will transfer the Pro-AG framework and novel integrated phage-based systems to several bacterial strains of concern to aquaculture with the goal of diminishing their antibiotic resistance (AR) genes and virulence potential.
What is a Focused Research Organization?
Focused Research Organizations (FROs) are time-limited mission-focused research teams organized like a startup to tackle a specific mid-scale science or technology challenge. FRO projects seek to produce transformative new tools, technologies, processes, or datasets that serve as public goods, creating new capabilities for the research community with the goal of accelerating scientific and technological progress more broadly. Crucially, FRO projects are those that often fall between the cracks left by existing research funding sources due to conflicting incentives, processes, mission, or culture. There are likely a large range of project concepts for which agencies could leverage FRO-style entities to achieve their mission and advance scientific progress.
This project is suited for a FRO-style approach for three reasons. First, it would be very difficult to attract VC or industry funding for this effort. The expected timeline is too long for most VCs who want to see a shorter horizon on return for their investments (on the order of 2-3 years). Second, the project has significant technical risk since we do not know how the Pro-AG systems will perform in the context of large enclosures densely packed with fish, which is a daunting environment for any anti-microbial intervention. Third, the scale of just the laboratory component of the project exceeds the level of funding normally available through standard channels of support for academic science, since Pro-AG delivery systems would need to be engineered in parallel for several different species of fish pathogens. This will also require more “applied” work than is typically supported by many academic research programs. For these reasons, the project fits perfectly in the sweet spot for a FRO.
How This Project Will Benefit Scientific Progress
If successful, our systems would greatly reduce the necessary frequency and concentrations of antibiotics to control bacterial fish pathogens. Solving or attenuating this central challenge to land-based aquaculture should help foster safe, sustainable and affordable sources of nutritious, uncontaminated fresh fish and help catalyze a shift away from unsustainable overfishing practices in the open ocean and environmentally hazardous practices in ocean-based aquafarms. This project could also have broader knock-on effects by enabling similar technical advances to reduce antibiotic resistance prevalence in other environmental settings (e.g., livestock, sewage treatment), which are also substantial sources of worldwide antibiotic resistance.
Key Contacts
Author
- Dr. Ethan Bier, UCSD, ebier@ucsd.edu
Referrers
- Alice Wu, Federation of American Scientists, awu@fas.org.
Learn more about FROs, and see our full library of FRO project proposals here.
A Focused Research Organization to Develop RNA Sequencing Technologies
RNA therapeutics are gaining popularity since they are cheap and easy to make, but sequencing technologies today rely on converting RNA back to cDNA, which collapses information on the more than 150 different chemically modified bases for RNA into just four bases. To address this knowledge gap, this project aims to develop direct sequencing tools specifically for RNA including chemical modifications in order to enable the complete sequencing of RNA bases and improve RNA therapeutics.
RNA encodes regulatory information that directs cellular functions. This dynamic information is written in four canonical bases, each of which can be chemically modified to create more than 150 different bases. Today, sequences of RNA are mostly obtained by converting RNA back to cDNA which is then sequenced; in that process of reverse transcription, all the modifications on RNA are lost. This knowledge gap significantly weakens our understanding and treatment of human diseases.
Project Concept
As a Focused Research Organization, this project will recruit an interdisciplinary team to develop a low-cost tabletop device that can sequence RNA, including chemical modifications, as accurately as we sequence DNA, and make this product available to researchers at large. We propose to 1) improve high-resolution imaging technologies, 2) develop single-cell mass spectrometry for RNA sequencing, and 3) advance nanopore technology to enhance their accuracy and resolution. Ultimately, the imaging technology will be coupled with the mass spectrometry or nanopore technology to extract full-length RNA transcripts from subcellular locations and then sequence them. In parallel, we will develop algorithms to deconvolute data and build data displays to quantitatively reflect the locations of RNA transcripts and their sequences with all modifications.
What is a Focused Research Organization?
Focused Research Organizations (FROs) are time-limited mission-focused research teams organized like a startup to tackle a specific mid-scale science or technology challenge. FRO projects seek to produce transformative new tools, technologies, processes, or datasets that serve as public goods, creating new capabilities for the research community with the goal of accelerating scientific and technological progress more broadly. Crucially, FRO projects are those that often fall between the cracks left by existing research funding sources due to conflicting incentives, processes, mission, or culture. There are likely a large range of project concepts for which agencies could leverage FRO-style entities to achieve their mission and advance scientific progress.
This project is suited for a FRO-style approach because academia is too siloed to support the kind of large-scale, interdisciplinary research project that this would require, and companies that currently provide DNA-based RNA sequencing tools have no motivation to innovate beyond tweaks to the current methods.
How This Project Will Benefit Scientific Progress
The development of direct RNA sequencing technologies with all chemical modifications will enable the execution of the Human RNome Project, which is anticipated to result from the the National Academies of Sciences, Engineering and Medicine Report (NASEM) on RNA Sequencing. A similar NASEM report resulted in the creation of the Human Genome Project. The Human RNome Project aims to identify all the possible chemical modifications of RNA and create a true sequence of RNA, i.e. the RNome. Direct RNA sequencing technologies will also enable scientists to sequence the complete genome of RNA viruses, such as SARS-CoV-2, the hepatitis C virus, and influenza viruses and accelerate the development of new RNA therapeutics for these viruses and other diseases.
Key Contacts
Authors
- Vivian Cheung, University of Michigan, Life Sciences Institute, vgcheung@umich.edu
- George Langford, Syracuse University
Referrers
- Alice Wu, Federation of American Scientists, awu@fas.org
Learn more about FROs, and see our full library of FRO project proposals here.
A Focused Research Organization to Design and Synthesize Spiroligomer Catalysts
This FRO will design and synthesize a library of spiroligomer enzyme-like catalysts to enable the development of new industrial processes for the production of green fuels and chemicals.
Humanity needs catalysts to create fuel, feedstocks to make materials, and fertilizers to grow food. Catalysts allow us to arrange atoms into the molecules we need with extremely high selectivity, cleanliness, and low energy input. Ever since Emil Fischer first conceived of the “lock and key” hypothesis of enzyme function, scientists have dreamed of rationally designing enzyme-like molecules. In 2021 the Nobel prize was awarded to Benjamin List and David MacMillan for developing organocatalysis – organic molecules that demonstrate basic catalytic function in enzymes. However, organocatalysts demonstrate only a small fraction (1/1,000,000,000) of the natural activity of the most capable enzymes because they are too small and do not display the deep, complex pockets of enzyme active sites needed to stabilize the transition states of reactions.
Project Concept
Spiroligomer nanostructures enable the creation of deep, complex, structured pockets that will allow us to design, assemble, and understand much more capable active sites. Using spiroligomer synthesis technology developed and scaled up over the last three years, this FRO will design, synthesize, and screen a library of spiroligomer catalysts. This will require the additional work of X-ray structure determination of active catalysts, measurement of activity using chemical kinetics, and computational modeling of active sites and reaction transition states.
What is a Focused Research Organization?
Focused Research Organizations (FROs) are time-limited mission-focused research teams organized like a startup to tackle a specific mid-scale science or technology challenge. FRO projects seek to produce transformative new tools, technologies, processes, or datasets that serve as public goods, creating new capabilities for the research community with the goal of accelerating scientific and technological progress more broadly. Crucially, FRO projects are those that often fall between the cracks left by existing research funding sources due to conflicting incentives, processes, mission, or culture. There are likely a large range of project concepts for which agencies could leverage FRO-style entities to achieve their mission and advance scientific progress.
This project is suited for a FRO-style approach because…Developing enzyme-like catalysts requires the engineering of highly structured molecules that are at least ten times larger than the kinds of molecules that synthetic chemists commonly create (5,000 Daltons for enzyme-like catalysts rather than 500 Daltons for typical small molecule therapeutics or organocatalysts). This will require a large engineering team with complex automation, instrumentation, and computation capabilities and professional synthetic chemists.
How This Project Will Benefit Scientific Progress
Unlike natural proteins that unfold and lose their activity when removed from their optimal temperature and solvent conditions, the spiroligomer-based catalysts we develop will be much more robust and valuable as industrial catalysis. Using synthetic chemistry, they can be produced at scale and made with much better quality control than natural proteins. It will also be easier to modify and tune their properties to create desired products. These catalysts display a much wider variety of chemically reactive groups than proteins do. This will open up the possibility of entirely new chemical production processes, such as artificial photosynthesis to create clean fuels and the production of other green chemicals and feedstocks.
Key Contacts
Author
- Christian Schafmeister, Temple University, meister@temple.edu
Referrers
- Alice Wu, Federation of American Scientists, awu@fas.org
Learn more about FROs, and see our full library of FRO project proposals here.
A Focused Research Organization to Characterize Antibodies Through Open Science
Many antibodies that scientists purchase from commercial manufacturers to conduct their research do not work as advertised, because most have never been validated properly. This project brings together the public and private sectors to conduct independent, third-party testing of commercial antibody manufacturers’ catalogs and publish the results in the public domain, such that no scientist ever uses an ineffective antibody again.
Thousands of scientists use antibodies – each of which targets one of the 20,000 human proteins – to develop fundamental theories of human biology, and to identify targets for new medicines. These antibodies are often purchased from commercial antibody manufacturers, whose combined catalog contains between 3.5 million and 4.8 million products. But for more than 30 years, the scientific community has been aware that many of these antibodies do not work as advertised, meaning that they do not recognize the intended protein target, or recognize the target but also recognize non-specific targets that confound their use. This occurs because many if not most antibodies have never been validated, or have been validated using inferior or outdated scientific methods, and because academics do not have resources or skill sets to test them themselves. When an antibody binds to a non-targeted protein, a researcher may believe that the target protein, perhaps a drug target, is present in a particular cell type or subcellular organelle when in reality it is not. These erroneous results lead to a vast waste of time, resources, and human capital.
Project Concept
The science on the optimal antibody testing methodology is largely settled: using an appropriately selected wild type human cell and a CRISPR knockout version of the same cell as the basis for testing yields the most rigorous and broadly applicable results. However, the cost of testing for an individual target or antibody is often prohibitive for any individual academic lab or company. Our organization, YCharOS (Antibody Characterization through Open Science), couples the settled science with a unique open science business model, in which a consortium of antibody manufacturers provide, in-kind, all their renewable antibodies (i.e. monoclonal or recombinant, which once tested are of value in perpetuity) to any given target to YCharOS for use in direct, head-to-head comparisons. This centralized testing model creates massive economic efficiencies for the sector while also providing immense scientific benefit to the public. Moreover, since all data will be released into the public domain using the principles of open science, the benefits accrue to all. We envision a world where no scientist ever uses an antibody that has not been rigorously tested by an independent third party. We believe that renewable antibodies for all 20,000 human proteins can be knockout validated in many applications for a one-time total budget of approximately $100 million.
What is a Focused Research Organization?
Focused Research Organizations (FROs) are time-limited mission-focused research teams organized like a startup to tackle a specific mid-scale science or technology challenge. FRO projects seek to produce transformative new tools, technologies, processes, or datasets that serve as public goods, creating new capabilities for the research community with the goal of accelerating scientific and technological progress more broadly. Crucially, FRO projects are those that often fall between the cracks left by existing research funding sources due to conflicting incentives, processes, mission, or culture. There are likely a large range of project concepts for which agencies could leverage FRO-style entities to achieve their mission and advance scientific progress.
This project is suited for a FRO-style approach because antibody characterization is a time limited project that, once completed, will identify high-performing antibodies that can be produced and used in perpetuity. Antibody validation itself is unlikely to generate papers, but will create a public good that enables the production of new research results using properly validated antibodies.
How This Project Will Benefit Scientific Progress
Academic and pharmaceutical scientists laboring to advance our understanding and treatment of human disease will be able to save time and money and produce higher quality research using validated antibodies. Monetarily, scientists spend an estimated $1 billion per year on ineffective antibodies that could otherwise be spent on conducting further research. Furthermore, there is a not insignificant volume of faulty research publications that have resulted from scientists unknowingly using ineffective antibodies.
Key Contacts
Authors
- Chetan Raina, CEO of YCharOS, chetan.raina@ycharos.com
- Al Edwards, CEO of the Structural Genomics Consortium, aled.edwards@utoronto.ca
- Peter McPherson, McGill University, peter.mcpherson@mcgill.ca
Referrers
- Alice Wu, Federation of American Scientists, awu@fas.org
Learn more about FROs, and see our full library of FRO project proposals here.
A Focused Research Organization to Quantify Ocean Carbon
The [C]Worthy Project will create first-of-its-kind, open-source software infrastructure for ocean carbon measurement, reporting and verification (MRV) to help drive the nascent marine-based carbon dioxide removal market.
There is scientific consensus that industrial-scale Carbon Dioxide Removal (CDR) will be necessary to meet the Paris Agreement’s goal of keeping the rise in global temperature to within 1.5°C or 2°C. By enhancing the ocean’s natural capacity for carbon sequestration and long-term storage, ocean-based CDR is one of the few strategies with the potential to remove carbon at the necessary scales. While investments are pouring into the design and early-stage deployment of ocean-based CDR technologies, the ocean is a complex ecosystem, a constantly moving fluid, posing challenges to quantifying ocean-based CDR projects. It is essential to establish scientifically credible methods for quantifying net carbon removal from CDR deployments and codify these as standards for Monitoring, Reporting, and Verification (MRV). [C]worthy is proposing transformative, open-source, technical solutions to confront these challenges.
Project Concept
[C]worthy aims at building the first foundational open-source computational infrastructure for MRV of ocean-based carbon dioxide removal technologies. The [C]worthy team will develop an innovative and first-of-its-kind MRV software infrastructure applicable across the range of ocean-based CDR technologies that are currently under study and testing in the US and internationally. This MRV platform will integrate ocean observations and advanced Earth system modeling tools at regional to global scales; it will incorporate ocean biogeochemistry and ecosystem models and data assimilation capabilities and it will advance techniques for efficient computation and analysis. This platform will be designed as open-source infrastructure with interoperable components—ensuring transparency and broad access to a growing CDR community.
What is a Focused Research Organization?
Focused Research Organizations (FROs) are time-limited mission-focused research teams organized like a startup to tackle a specific mid-scale science or technology challenge. FRO projects seek to produce transformative new tools, technologies, processes, or datasets that serve as public goods, creating new capabilities for the research community with the goal of accelerating scientific and technological progress more broadly. Crucially, FRO projects are those that often fall between the cracks left by existing research funding sources due to conflicting incentives, processes, mission, or culture. There are likely a large range of project concepts for which agencies could leverage FRO-style entities to achieve their mission and advance scientific progress.
This project is suited for a FRO-style approach because the [C]worthy development project requires a level of coordinated computational engineering that is too big for a single academic lab, too complex for a loose multi-lab collaboration, and not directly profitable enough for a venture-backed startup or industrial R&D project. It is aiming to develop an open-source IT infrastructure, which is a product of public benefit for science and technology. This requires tight-knit coordinated effort in a fast-paced start-up-like environment. It is therefore best fit for the FRO model.
Key Contacts
Team lead
- Matt Long, PhD, matt@cworthy.org
Referrers
- Alice Wu, Federation of American Scientists, awu@fas.org
Learn more about FROs, and see our full library of FRO project proposals here.
A Focused Research Organization to Build the Foodome Project for the Future of Nutrition
Our current knowledge of the biochemical compounds in food is incredibly limited, but existing databases of MassSpec scans contain massive amounts of untapped, unannotated information about food ingredients. A project to leverage these databases with the tools of data mining, AI, and high-throughput measurement will systematically unveil the chemical composition of all food ingredients and revolutionize our understanding of food and health.
Diet is the single biggest determinant of health over which we have direct control. An unhealthy diet poses more risk to morbidity than alcohol, tobacco, drug use, and unsafe sex combined. Indeed, our diet exposes us to thousands of food molecules, many of which are known to play an important role in multiple diseases including coronary heart disease, cancer, stroke, and diabetes. Despite the demonstrated and complex role of diet on health, nutrition science remains focused on molecules that serve as energy sources such as sugars, fats, and vitamins, leaving most disease-causing compounds uncatalogued and invisible to researchers and health care professionals. Further, our current understanding of the way food affects health is limited to nutritional guidelines that rely on a panel of 150 essential micro- and macro-nutrients in our diet. This is a tiny fraction of the more than 130,000 compounds known to be present in food, hence limiting our ability to unveil the health implications of our diet.
Project Concept
The Foodome project aims to unveil this “dark matter of nutrition” by creating an open-access high-resolution compendium of food compounds through a strategy that combines Big Data, ML/AI, and experimental techniques, implemented by a focused cross-disciplinary team, motivated to bring transformative change and maximize public benefits.
In the past five years, BarabásiLab has curated the largest library of compounds in food, consisting of more than 135,000 biochemicals linked to 3,500 foods. While the number of biochemicals is exceptional, the coverage is highly uneven, sparse, and largely unquantified. Yet, information about the missing biochemicals is carried by the unannotated MassSpec peaks available for each MassSpec scan of food ingredients. Because chemicals are invisible to the one-chemical-one-peak tools employed today, we have designed a strategy that relies on data mining, AI, and high-throughput measurements to resolve them: we plan to collect the more than 3,000,000 MassSpec scans already available in databases, and mine the full scientific literature to collect knowledge on food composition. We also plan to take advantage of the increasing number of annotated genomes to infer their chemical makeup. These data will serve as input for a ML/AI platform designed to learn associations between biochemical structures and the ingredients’ phylogenetic position, helping us systematically unveil the chemical composition of all food ingredients.
What is a Focused Research Organization?
Focused Research Organizations (FROs) are time-limited mission-focused research teams organized like a startup to tackle a specific mid-scale science or technology challenge. FRO projects seek to produce transformative new tools, technologies, processes, or datasets that serve as public goods, creating new capabilities for the research community with the goal of accelerating scientific and technological progress more broadly. Crucially, FRO projects are those that often fall between the cracks left by existing research funding sources due to conflicting incentives, processes, mission, or culture. There are likely a large range of project concepts for which agencies could leverage FRO-style entities to achieve their mission and advance scientific progress.
This project is suited for a FRO-style approach because the Foodome platform and knowledge base will address problems in health science beyond the competence of any single academic group or start-up. The project started in the academic environment involving groups at Northeastern University, Harvard Medical School, and Tufts Medical School, but typical academic researchers and institutions are motivated by short term publication strategies and unable to devote the years needed to develop a public resource. Federal nutrition research funding exists, but is fragmented, and normal funding channels are generally unable to offer sustained support for a project of this size. With VC funding, we were able to move the project to a startup environment to standardize the toolset and develop key technologies, but company management decided that the Foodome platform’s timeline is too far from the market. Based on these experiences, an FRO appears to be the best framework to accomplish the vision of Foodome. The project enters a field limited by technological stagnation, and will fundamentally change our understanding of health and disease, impacting multiple fields and industries.
How This Project Will Benefit Scientific Progress
A high-resolution knowledgebase on the composition of food will revolutionize our ability to explore the role of each food-borne molecule in human health, impacting multiple fields: 1) It will be transformative for health care, changing our ability to prevent and control disease. 2) It will aid the development of healthier, more nutritious, and biochemically balanced foods. 3) It will facilitate the development of novel pharmaceuticals. 4) By improving MassSpec annotations, it will provide a more accurate biochemical descriptions of any sample, empowering diagnosis, and detection. 5) It will unlock innovations in personalized nutrition and precision medicine, allowing clinicians to offer precision advice to a patient on how to use diet to prevent and manage disease.
Key Contacts
Author
- Albert-László Barabási, Northeastern and Harvard Medical School, alb@neu.edu
Referrers
- Alice Wu, Federation of American Scientists, awu@fas.org
Learn more about FROs, and see our full library of FRO project proposals here.
A Focused Research Organization for Superconducting Optoelectronic Intelligence
Artificial intelligence places strenuous demands on current computing hardware, but the integration of semiconductors, superconductors, and optical hardware will create revolutionary new tools for AI.
Digital computers are excellent for number crunching, but their operations and architecture contrast with the operations that support intelligence. When used for AI, vast amounts of energy, data, and time are required to train new models. Meanwhile, the field of computational neuroscience relies on these digital computers to simulate cognition. Because the underlying computational hardware is poorly matched to the operations of synapses, dendrites, and neurons, the same problems with time and energy arise. We can address both these needs with advances in computer hardware.
Project Concept
We can address both these needs with advances in computer hardware. Our approach builds upon the silicon transistors of digital computing, adding superconducting circuitry to accomplish neural computations, and optical components to realize extensive communication across human-brain-scale systems. We have already made substantial progress in demonstrating key components and are ready to scale to a multiyear effort to integrate into a chip-scale cortex (see slides).
You can learn more about superconducting optoelectronic networks in this slide deck.
What is a Focused Research Organization?
Focused Research Organizations (FROs) are time-limited mission-focused research teams organized like a startup to tackle a specific mid-scale science or technology challenge. FRO projects seek to produce transformative new tools, technologies, processes, or datasets that serve as public goods, creating new capabilities for the research community with the goal of accelerating scientific and technological progress more broadly. Crucially, FRO projects are those that often fall between the cracks left by existing research funding sources due to conflicting incentives, processes, mission, or culture. There are likely a large range of possible project concepts for which agencies could leverage FRO-style entities to achieve their mission and advance scientific progress.
This project is suited for a FRO-style approach because the integration of semiconductors, superconductors, and optical hardware is beyond the scope of a single academic or government research group, and this endeavor will require appreciable investment in a well-orchestrated, focused team, more akin to a startup. However, given the complexity of the technology, five years will be required to bring a competitive product to market, which is still too early for venture capitalists. Because AI hardware is well-established and continuously improving, gaining market traction will require not only superior hardware, but also streamlined software and user interfaces. An FRO is the ideal context to pursue a complete system meeting the needs of a large and diverse pool of users.
How This Project Will Benefit Scientific Progress
By realizing superconducting optoelectronic networks, we will achieve cognitive AI with vastly more computational power than has been possible with the largest supercomputing clusters of today, while consuming only a fraction of their power. Continued scaling of our technology will not come at the cost of environmental harm. Scientists, engineers, and entrepreneurs across the country will have access to a revolutionary new tool to interpret and analyze complex, multi-modal datasets. This form of advanced AI will change how we provide health care, harness energy, model Earth’s climate, and more. Superconducting optoelectronic hardware will usher the largest transition in computation since silicon, enabling powerful tools for computing and an experimental testbed to elucidate the mechanisms of our own minds.
Key Contacts
Author
- Dr. Jeffrey Shainline, NIST, jeffrey.shainline@nist.gov
Referrers
- Alice Wu, Federation of American Scientists, awu@fas.org
Learn more about FROs, and see our full library of FRO project proposals here.